New Features in PACE v4.0.0
A new release of PACE (Performance Analytics for Computational Experiments), the performance monitoring framework for E3SM has been deployed on April 25th, 2022. PACE automatically captures and stores the model provenance and timing information of every E3SM experiment executed on DOE supported supercomputers in a database and makes the performance charts and metadata accessible to E3SM users through a web portal (). The latest release 4.0.0 introduces the following major features. A researcher can search for an experiment of interest and access the following new performance plots from the experiment details page.
I/O Performance
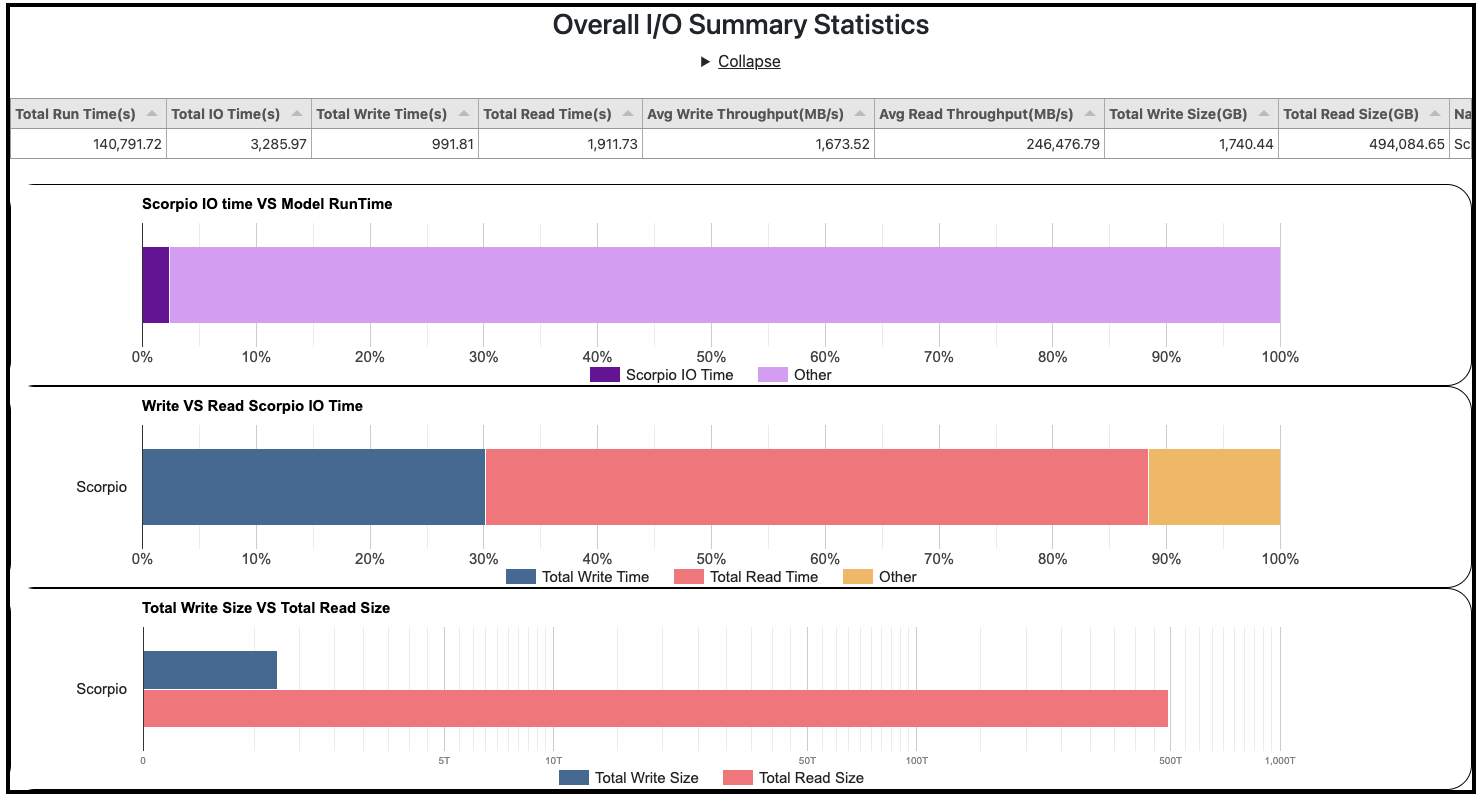
PACE now includes Input/Output (I/O) performance metrics in addition to detailed timing profiles that were already available. This capability has been added by the recent release of the Scorpio I/O library. The web portal presents an overall I/O performance summary (Fig. 1) for every experiment including the portion of time spent performing I/O relative to the total execution time of the experiment. Additionally, the size of data read/written, the time spent reading/writing data and the average throughput rates are provided at the application level.
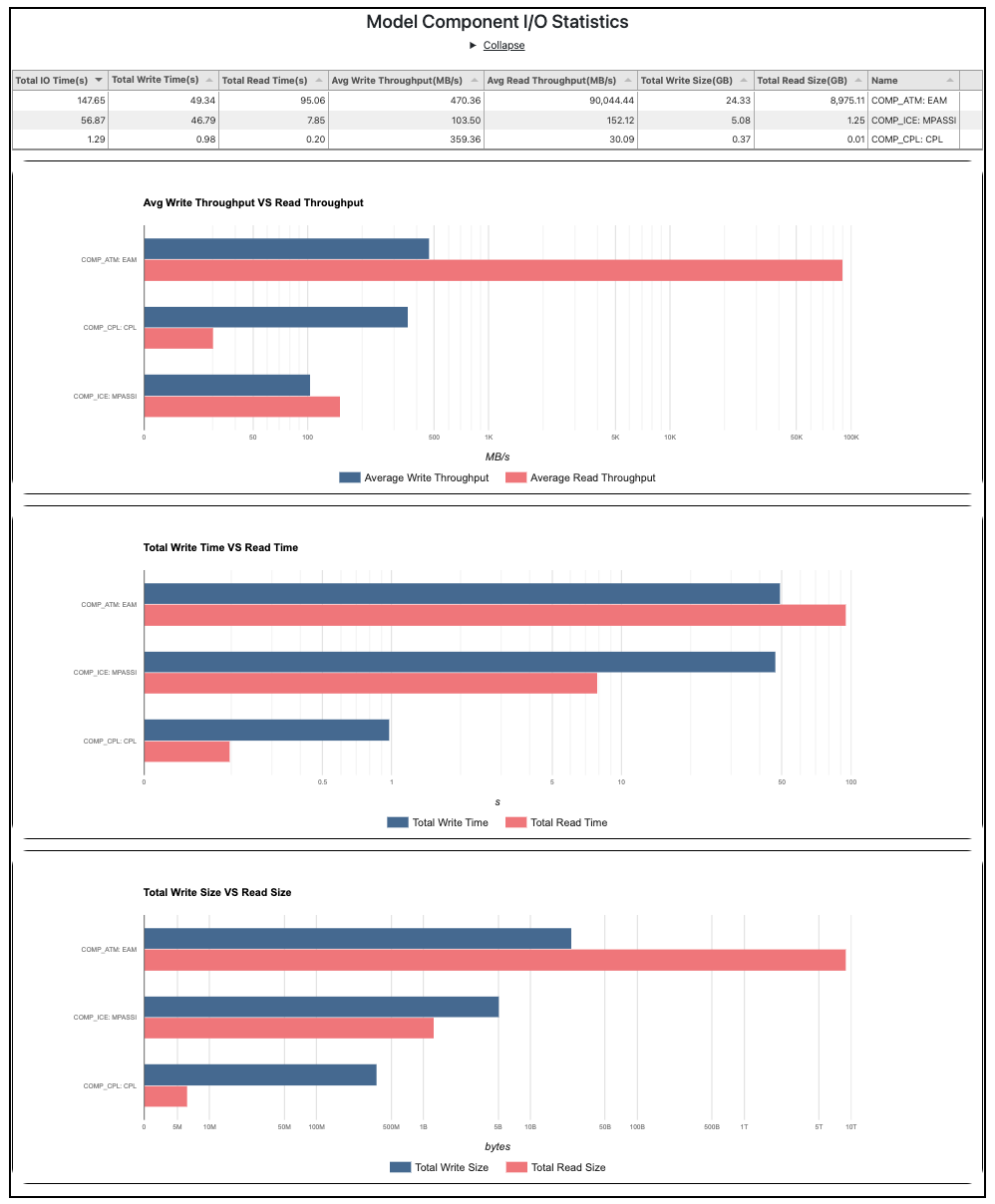
Furthermore, these detailed metrics (read/write times, sizes, average throughput rates, Fig. 2) are presented for each model component and at a finer granularity for each input/output file. This information can provide insights into costs of I/O and help identify bottlenecks that need to be addressed.
Memory Profiles
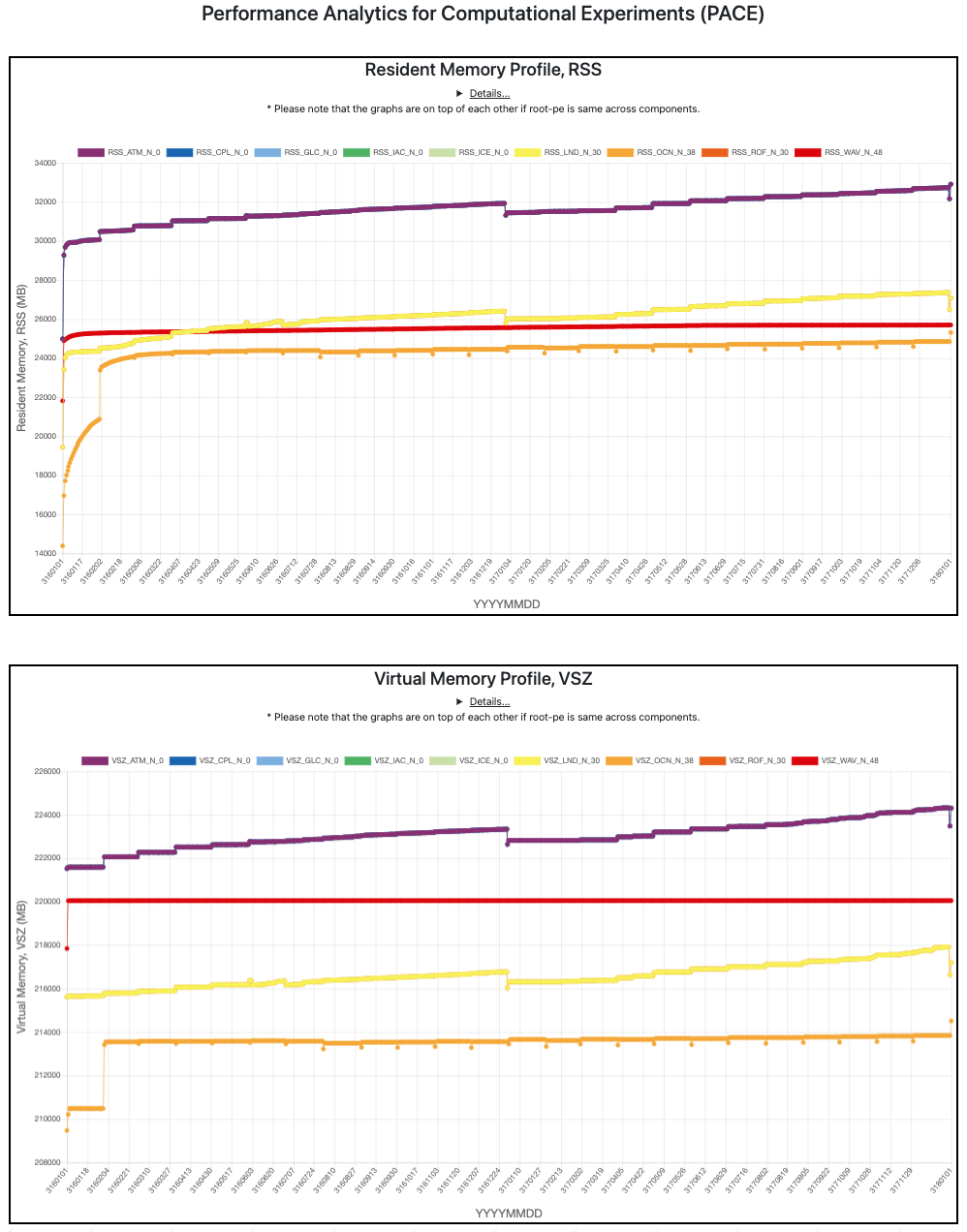
PACE web portal displays detailed memory usage of the model across various components during the course of execution (Fig 3.). These memory profiling plots can identify potential memory leaks as well as aid in tracking down out of memory errors.
Build Times
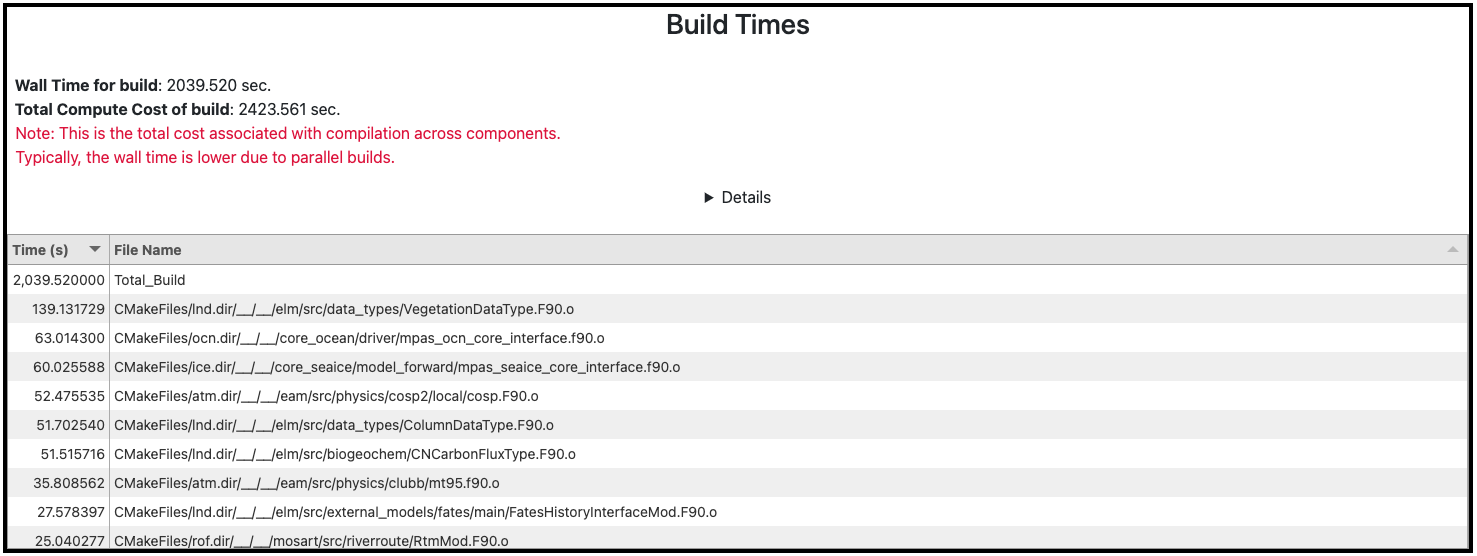
PACE now provides detailed information of the compilation costs associated with building the model for a particular configuration (Fig. 4). This information can assist in addressing excessively long compilation times by identifying problematic files so that a researcher can take remedial actions such as reducing compiler optimization level or turning off specific optimizations.
Back End Improvements
Additionally, this PACE release included a significant redesign and refactoring of the parser backend with a better separation of concerns that has already facilitated extensibility and will improve future maintainability. This work has laid the foundation for incorporating all these new capabilities.
Finally, we have also surpassed a noteworthy milestone of archiving 100,000 E3SM experiments in PACE.
References
If you are not familiar with other performance summaries and plots available from PACE, please check the post Performance Analytics for Computational Experiments (PACE) with detailed explanations.
- Main page on e3sm.org: PACE
- PACE portal: PACE – Home
- Videos:
- PACE documentation: https://pace-docs.readthedocs.io/
- News post, 2019/11: Performance Analytics for Computational Experiments (PACE)
Funding
This research was supported as part of the E3SM project, funded by the U.S. Department of Energy, Office of Science, Office of Biological and Environmental Research.
Acknowledgments
Thanks to Jayesh Krishna, Azamat Mametjanov and Jim Foucar for their assistance in incorporating these new capabilities.
Contact
- Gaurab KC and Sarat Sreepathi, Oak Ridge National Laboratory
This article is a part of the E3SM “Floating Points” Newsletter, to read the full Newsletter check: